|
Do not use the computer in any way to do this exercise.
-
Find the best global alignment between the DNA sequences:
AGTAGTCAAG
AGAAGTAAG
Use the following scoring system:
-
Find the best local alignment between amino acid sequences:
using the BLOSUM 62 score matrix (which can be found in the notes of
the 3/6 lecture) and a gap penalty w(k) = -1 - 2k.
Solution:
To arrive at the best local
alignment the following matrix was constructed:
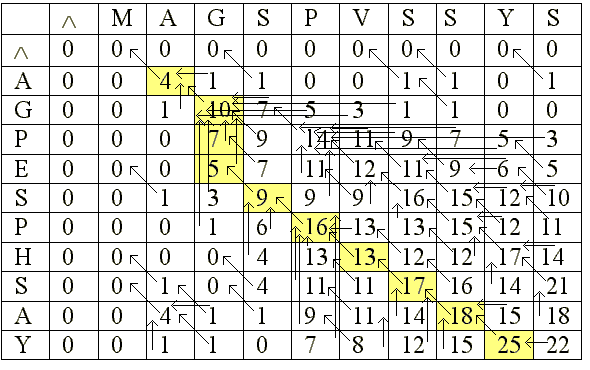
The scores entered in the cells of this matrix were derived similarly
to the method described for the global alignment with a few
exceptions: any score resulting in a negative score is given a score
of 0; scores for matches and mismatches are determined by referring to
the BLOSUM 62 scoring matrix.
To determine the final alignment, first find the cell with the highest
score in the matrix. Follow the arrows until a 0 is reached. This
will give the best local alignment.
|